Sub-theme C
Bridging basic medicine and clinical medicine by fusing heart simulation and molecular simulation
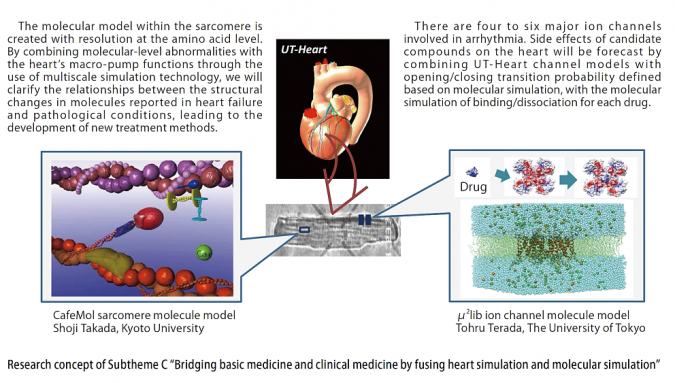
In development activities by UT-Heart so far through the use of the K computer, multiscale simulation technology connecting micro contraction proteins which constitute the sarcomere with macro heartbeats was successfully developed.
For the post-K(Supercomputer Fugaku) project, we will take this technology further to develop a sarcomere model consisting of coarse-grained molecular simulation models, and coordinate with the heart model. We consider that this will enable us to, for example, understand how the load observed and measured at the macro level is conveyed to the molecular level and activates the intracellular signal transducing system, and clarify the mechanism for causing pathological conditions. Moreover, by using the molecular model, we will build the ion channel which controls the input/output into/from the cells of various ion currents which contain calcium ions that modulate the motion of contraction proteins, and simulate binding/dissociation to/from drugs, to evaluate the risk of arrhythmia caused by drugs to the heart. This will enable swift screening of candidate compounds which produce side effects on the heart and make the process of drug design more efficient. This study will be conducted in tandem with Priority issue 1.
Group Leader
Toshiaki HISADA (UT-Heart Inc.)
Research concept of Sub-theme C
Achievements
(Japanese Only)